Addressing key questions in evolution
How and when species came to be is the fundamental question in macroevolution. Attempts to answer it use a variety of data sources including genome sequences, morphology, and fossil discoveries. Currently, there is no method for exploiting all of this data simultaneously, while analysing different data sources individually often produces conflicting results.
At the University of Auckland’ s Centre for Computational Evolution, a research team led by Professor Alexei Drummond, Dr David Welch and Dr Nick Matzke are using NeSI computing resources to try and address this challenge.
They are developing new mathematical models that will combine genomic, fossil and phenotypic data from multiple sources to give us the best possible understanding of evolutionary history.
“The methods we develop to analyse data are very computationally intensive,” says Walter Xie, a research programmer at the Centre. “To analyse a single data set under one set of assumptions may require days for computing time on a regular machine. We typically analyse data under multiple scenarios and need to check and double check that the analysis is valid, so we need a resource like NeSI to be able to use many machines in parallel.”
In fact, NeSI resources are an essential tool even before the researchers get to their analysis stage.
“A large part of our work is developing new methods to analyse multiple forms of data in a combined analysis,” Walter says. “Since we are developing new methods, we need to validate them on multiple simulated datasets before we can even start using them to analyse real data.”
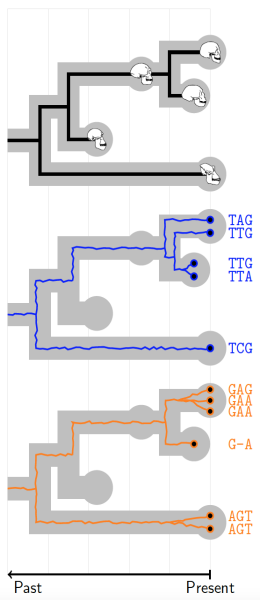
A species tree displaying ancestral relationships between modern and ancient species (top). To reconstruct this tree we have morphological data from fossils and modern samples and multilocus sequence data from recent samples only (middle and bottom). A unified model accounts for all these data simultaneously.
There are two important steps in their work. First, they need to demonstrate that the method works and produces the correct result under the assumptions of the model. This is called model validation. A typical validation involves simulating data sets under a model with known parameter values, then using their methods to see if they can recover the correct parameter values.
“NeSI is used both to simulate the data and run the subsequent analysis,” Walter explains. “The analysis of a simulated data set may take hours to run and 100-500 simulated data sets may be analysed. This whole process is repeated many times as the model is refined and debugged.”
Once they are satisfied their methods are correct, they can then apply it to real data. They will analyse data sets of interest under various models and assumptions, and use model selection tools to compare them and find the most robust answer. This often comes with its own set of challenges.
“Real data is typically much more difficult to analyse than simulated data,” says Walter. “For a start, we don’t know the answer before we begin. The methods and models usually require lots of tweaking before reliable results start to appear.”
The final output of an analysis is typically a phylogenetic tree that displays the ancestral relationships between the species being analysed and the estimated values of parameters of the model, for example, fossilisation rates over the epochs.
“Without NeSI, these computationally intensive studies would simply not be viable,” says Walter. “The batch processing using NeSI saves a lot of our time, and the parallel computing power in NeSI is another big benefit for us.”
Looking ahead, Postdoc Fábio Kuriki Mendes is looking to expand their model’s capabilities even further, to better accommodate sampling bias in fossil samples. They will continue to use NeSI resources for their testing work.
More information on the project, which is supported by Marsden funding, can be found here. Outside of its New Zealand collaborators, the project also engages overseas experts Associate Professor Tanja Stadler and Dr Tim Vaughan from ETH Zurich, Dr. Mana Dembo and Dr. Mark Collard from Simon Fraser University, and Dr. Graham Slater from the University of Chicago.
---------------------
Do you have an example of how NeSI platforms have supported your work? We’re always looking for projects to feature as a case study. Get in touch by emailing support@nesi.org.nz.







