Understanding environmental processes on a microbial level
When 700 million litres of oil spilled into the Gulf of Mexico in 2010 from an oil tanker accident, it made headlines around the world for its impacts on the local environment and policies around drilling practices. Thankfully a major spill like that doesn’t happen every day, but studying the 2010 Deepwater Horizon accident has helped inform important research in NZ related to waterways pollution.
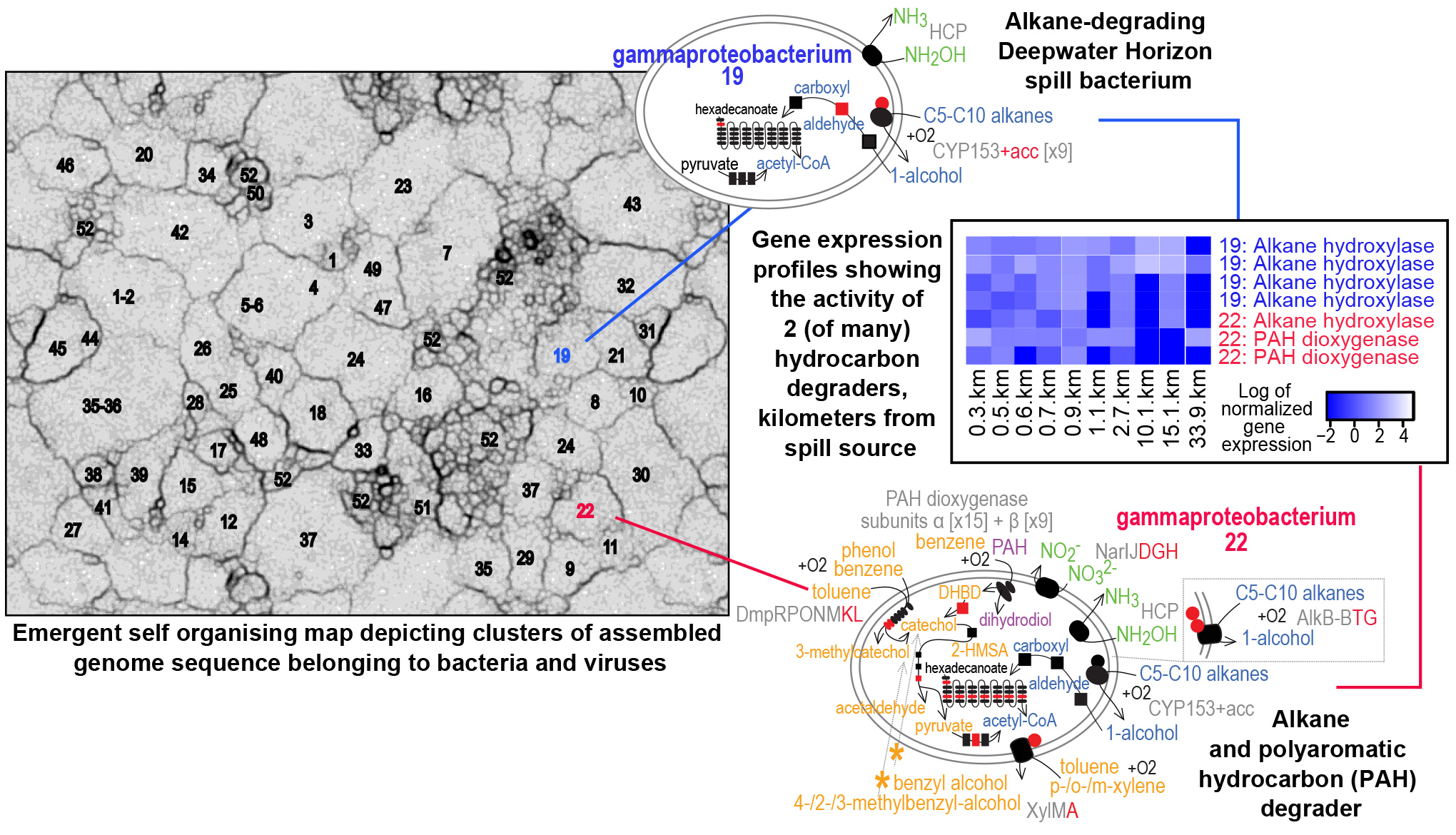
Dr. Kim Handley is a Senior Research Fellow in Biological Sciences at the University of Auckland. In one of her latest projects, she and colleagues used NeSI computing resources to analyse the genomes of 57 uncultivated bacteria they reconstructed from the Deepwater Horizon disaster’s post-spill deep-sea sediments and to study their gene expression patterns across the seafloor. Analyses from the study, published in the ISME Journal, suggest that a broad metabolic capacity to respond to oil inputs exists across a large array of usually rare indigenous deep-sea bacteria. Understanding these bacteria and their interactions with the oil could have valuable applications elsewhere.
“The aim of our group’s research is to link microbiology with environmental processes, such as the removal of pollutants from waterways,” Handley says. “To do this, we use approaches that enable us to study the genetic and metabolic profiles of microbial communities.”
Funded by a Royal Society of NZ Rutherford Discovery Fellowship, Handley is involved in a range of projects, from investigating the implications of sediment inputs on estuary function to studying how toxin-producing stream bacteria respond to nitrate pollution.
Access to NeSI’s high performance computing resources is integral to her group’s bioinformatic work. Studying microbial communities involves assembling dozens of microbial genomes at a time from large environmental datasets, working with massive output files, some of them hundreds of gigabytes each in size, and manipulating large search databases.
NeSI’s large memory machines are used for her group’s genome assemblies and assembly post-processing work, such as the assignment of assembled fragments to particular taxa, pairwise genome comparisons, and gene functional annotations.
“We’ve used NeSI to run arrays of hundreds of genome-related jobs in order to achieve rapid simultaneous processing,” Handley says. “In addition, use of a shared, remote computing resource enables the team to access data, databases, and software without redundancy, and from anywhere at any time.”
Technical staff at NeSI and Auckland’s Centre for eResearch have helped Handley and her team make the most of the supercomputing resources. In addition to providing standard support like assisting with software installs or troubleshooting occasional job errors, Martin Feller and Markus Binsteiner at the Centre for eResearch and Ben Roberts at NeSI also helped Handley and her students with large data transfers and provisioned custom spaces on NeSI’s Pan cluster for databases and software installations that were shareable across all projects.
“NeSI support have also assisted in working with local university IT to provide access to additional resources for data storage and machines for graphical user interface software and larger genome assemblies beyond Pan’s capacity,” she said.
Looking ahead, Handley has other projects on the horizon that will make use of NeSI resources, including an MBIE-funded project studying the genomic content of aquifers to understand groundwater as a living and reactive system, and a study that’s part of Genomics Aotearoa, the national advanced genomics platform, that looks at high quality genomes from waterways. For these and future projects, NeSI resources will remain an essential component of their work, she says.
“High performance computing is essential for researchers working with large and complex genomic datasets. Researchers attempting to assemble multiple (potentially hundreds of) genomes from diverse sediment microbial communities, for example, need access to large memory machines,” she says. “With that in mind, we are looking forward to migrating to NeSI’s new and enhanced-capacity high performance computing platform this year.”
- - - - - -
Recent publications that used NeSI resources:
1. 2017 Handley KM, Piceno YM, Hu P, Tom LM, Mason OU, Andersen GL, Jansson JK, Gilbert JA. Metabolic and spatio-taxonomic response of uncultivated seafloor bacteria following the Deepwater Horizon oil spill. The ISME Journal 11: 2569-83.
2. 2017 Lear G, Lau K, Perchec A-M, Buckley HL, Case BS, Neale M, Fierer N, Leff JW, Handley KM, Lewis G. Following Rapoport's Rule: The geographic range and genome size of bacterial taxa decline at warmer latitudes. Environmental Microbiology 19: 3152-62.
Current NeSI projects, collaborators and group members:
Estuarine pollution tolerance and nutrient cycling (PhD students Katie Worrallo and Jian Boey, School of Biological Sciences, University of Auckland; collaborator Professor Simon Thrush, Institute of Marine Science, University of Auckland)
The development of toxic stream biofilms (PhD student Hwee Sze Tee; collaborator Dr. Susie Wood, Cawthron Institute)
Marine sponge symbionts (collaborator Dr. Laura Steindler and PhD student Ilia Burgsdorf, University of Haifa, Israel)
The metabolic capacity of groundwater communities (PhD students Olivia Mosley and Emilie Gios, School of Biological Sciences, University of Auckland; collaborators Dr. Louise Weaver and Murray Close, ESR and Dr. Chris Daughney, GNS)
Geographic effects on microbial genomes (collaborator Associate Professor Gavin Lear and PhD student Jieyun Wu, School of Biological Sciences, University of Auckland)
Gut microbiomes and autism (collaborator Associate Professor Mike Taylor and PhD student Giselle Wong, School of Biological Sciences, University of Auckland)
- - - - - -
Do you have an example of how NeSI platforms have supported your work? We’re always looking for projects to feature as a case study. Get in touch by emailing support@nesi.org.nz.