Speeding up the post-processing of a climate model data pipeline
The below case study shares some of the technical details and outcomes of the scientific and HPC-focused programming support provided to a research project through NeSI’s Consultancy Service.
This service supports projects across a range of domains, with an aim to lift researchers’ productivity, efficiency, and skills in research computing. If you are interested to learn more or apply for Consultancy support, visit our Consultancy Service page.
Research background
Climate models are notorious for producing vast amounts of output data. The research of NIWA Climate Scientist Jonny Williams underpins and supports the earth system modelling capability of New Zealand researchers and the wider Deep South National Science Challenge. Jonny has a post-processing pipeline that takes global surface temperature and other fields to compute area means and point values for different diagnostics and climate models. The climate models are started in the past (e.g. 1950) and continue until (e.g. 2100). Jonny's research provides a glimpse into the future, which will help prepare stakeholders take action or mitigate the effect of global warming.
Project challenge
The post-processing steps required as part of Jonny's work can take up to several weeks to complete. Therefore, the aim of this Consultancy project was to reduce the turnaround time and increase Jonny's research productivity.
What was done
Rose is a framework for managing and running meteorological suites. A tool (rosesnip) was written to split multi-model and multi-diagnostic Rose configuration files into 100s of smaller files, each handling a single tasks that can be executed in parallel. We leveraged the Cylc workflow manager to submit the jobs to the SLURM scheduler. The approach adopted in this project was to rely on existing tools (afterburner, Cylc) whenever possible. Our solution, which consists of two Python scripts, produces the same result as the original serial rose-script but within a much shorter time frame.
An important outcome of the project is that the turnaround time is no longer limited by the number of models or diagnostics but instead by the maximum number of parallel tasks that can run concurrently on our platform. Nevertheless, scalability will be limited by load balancing -- some tasks still take a significant amount of time which can cause other tasks to be in "wait" mode. The figure below shows the distribution of the execution time for the 528 tasks. All the tasks finished in 36,000 seconds while the cumulative execution time was 1.3 million seconds, yielding a speedup of 35 for a maximum number of concurrent tasks set to 40. This represents a parallel efficiency of 88%.
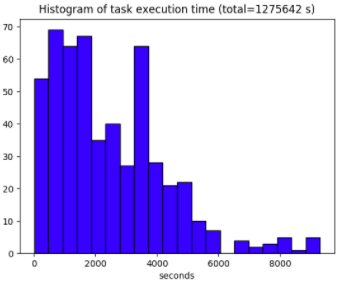
In contrast to other parallel programming models, waiting tasks don't block any resources, making the present approach more efficient than other technologies such as MPI or OpenMP.
The underlying software invokes the Iris Python module. We found that model output files take about one minute to load each (> 80% of execution time). The statistical computations on the other hand take less than 20% of the total execution time.
The Rosesnip scripts have been written with the goal of facilitating maintenance. Rosesnip does not have any external dependencies. The files produced by rosesnip are backwards compatible with those required by the underlying software (before NeSI’s involvement).
Main outcomes
- A significantly reduced turnaround time through task parallelisation. The computation of summary statistics can be performed independently and in parallel between models and across diagnostics. Given that there are 1000s of model and diagnostics combinations, the potential of speedup is significant.
- An improved utilisation of NeSI / NIWA HPC platforms. Instead of submitting a single job that sits in the queue for a very long time due to its wall clock requirements, the researcher can submit lots of smaller jobs, which can start quickly. These jobs can be monitored, paused, and restarted if desired.
Researcher feedback
"Thanks to NeSI I'm now running a case with around 85 diagnostics and 49 models which successfully caches its data as it goes along. Without this work, it would have taken months to run and is unlikely to have succeeded in one go anyway, since the machine requires periodic downtime and maintenance.
"Just to emphasise this point, before NeSI helped with this project, I was facing cases where I could not compute all the diagnostics required in a realistic time frame and therefore this consultancy has made possible what was simply impossible before.
"I am confident that this work will be of great interest to the UK Met Office who are the owners of the software upon which this consultancy was based. I am very grateful to NeSI for their support in this work and look forward to working together again."
- Jonny Williams, Climate Scientist, NIWA
Do you have an example of how NeSI support or platforms have supported your work? We’re always looking for projects to feature as a case study. Get in touch by emailing support@nesi.org.nz.







