Using HPC for Research and Career Development in Life Sciences
For many early career scientists, certain skills or experience gained through their projects and collaborations can be game changers in determining their future research path. In the case of Otago University PhD graduate Tom Kelly, one of his career turning points came via an introduction to programming and access to NeSI supercomputers.

Over the last four-and-a-half years, Kelly has used statistical approaches to study gene expression and genetic interactions, looking at the consequences of gene regulation for populations evolving and the dysregulation in cancers. Access to high performance computing (HPC) clusters, knowledge of the Linux interface, and use of programming languages such as R, became essential for gathering results for both his Honours and PhD projects.
“Drawing upon my mathematical skills and research interests, I've actively pursued research questions in computational biology that require statistical, computational, and HPC techniques to address,” he says. “Performing statistical tests across thousands of genes, and adjusting for multiple comparisons, can sometimes goes beyond the scope of what a local machine is capable of, especially when looking at interactions between pairs or combinations of genes.”
Whether he’s analysing biological pathways or testing a hypothesis where the underlying distribution is unknown, Kelly has come to rely on HPC to help him delve deeper into complex datasets.
“The role of HPC in assembling genomics is well known but there are other ways HPC is used in bioinformatics as well. I usually deal with processed data but the statistical procedures can also be computationally demanding,” he says. “It's been particularly fulfilling to gain sufficient scientific computing and HPC skills to be able to participate in eResearch conferences and have the expertise to teach programming workshops to students in the same situation as I was not so long ago.”
From time to time he called on Matt Healey and Peter Maxwell from NeSI’s Solutions Team, who were always quick to help with questions both big and small.
“The main thing that I appreciate is that they've been very patient with jobs that didn't work as intended,” says Kelly. “I've never hesitated to ask ‘dumb’ questions and it's been a great system to try things out and develop expertise. I'm increasingly aware that core-hours cost money and it's been a privilege to learn on the institutional access where this has not confined my learning opportunities or the research questions that I could pursue.”
With data analysis and computational techniques driving many of today’s research approaches, a greater need is emerging for researchers to build skills in these areas.
“We have the technology to generate vast amounts of genetic data and the computing resources to process it but very few people are equipped to handle it effectively,” Kelly says. “I support the Software Carpentry initiative wholeheartedly and am glad for the role NeSI has taken with it in New Zealand. I encourage anyone in the life sciences to take any opportunity to learn computational tools and see if it assists your research or suits you as a future career direction.”
Kelly recently completed a PhD project that focused on methods for addressing synthetic lethality in breast and stomach cancer. Synthetic lethality occurs when deficiencies - caused either by mutations, inhibitors or other factors - affects the expression of two more more genes and leads to the death of the cell.
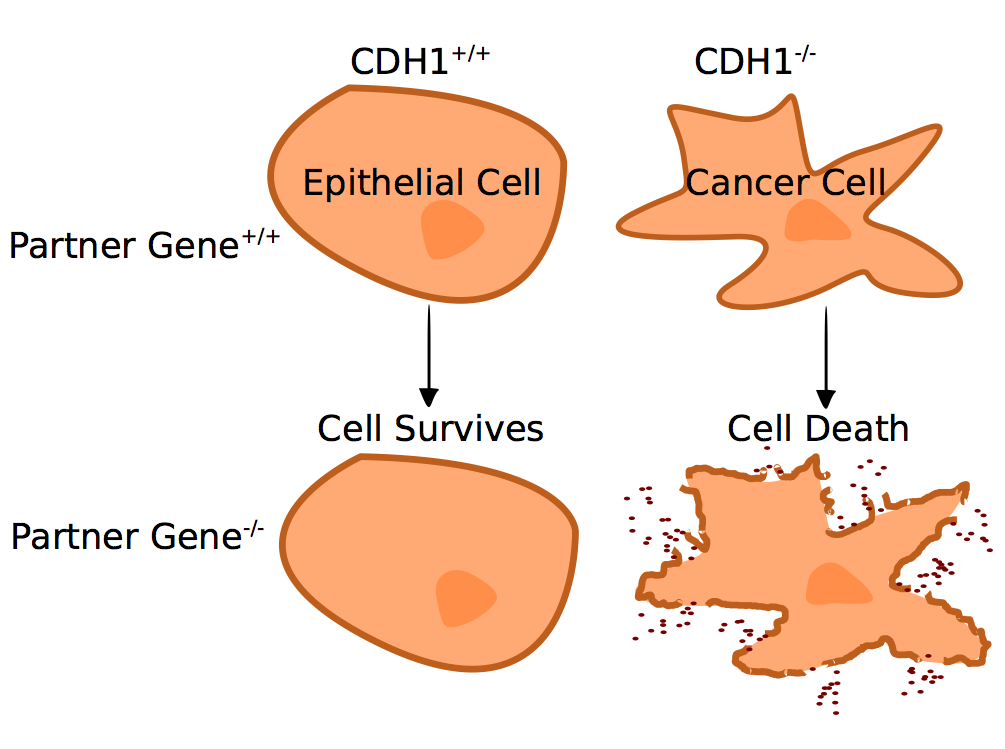
To address this, Kelly explored how to develop a bioinformatics tool to identify synthetic lethal partners of a (cancer) gene from expression data and design indirectly targeting medicines against genes lost in cancers.
His investigations were further extended to analyse and simulate synthetic lethal pathways to examine whether the method could be applied to molecular pathways in cancer. Access to NeSI has been essential yet again.
“These aren't as common in bioinformatics but the simulations, in particular, became a crucial part of my thesis to support the methods that I had already developed, applied, and released as R packages,” he says. “The simulations became a very computationally-intensive part of the project and are a lot stronger and more conclusive for the heavy use of HPC. Simulating large gene expression datasets and complex biological pathway networks required HPC to be performed and this aspect of the project was only pursued to this depth because of the skills I had developed in using R, Linux, and NeSI up to this point.”
With his PhD thesis now complete, Kelly has begun applying for postdoc positions in overseas, with a particular interest in a career in Japan. In Kobe, RIKEN operates the K supercomputer, named after the Japanese word "kei" (京) for the number 1016 and is currently ranked as the eighth-fastest computer in the world. Many research groups at the RIKEN research centres and Universities in Japan use the K supercomputer for their research. A postdoctoral position there would draw upon the programming and HPC experience Kelly gained with NeSI and provide opportunities to take his skills to yet another level.
Reflecting on how he has gotten to this point in his career, Kelly says his experience with HPC has been valuable for more than just the practical skills.
“I think the biggest thing that I have gained is experience tackling new problems with new technologies and having it pay off,” he says. “The opportunity to push myself in a new direction and see it come to fruition is why I pursued a career in science and using NeSI was an excellent way to include transferable skills and personal development in my PhD. HPC is definitely a useful resource to have access to and something I would encourage early career researchers to get some experience with. It will open up research questions that you would not have even considered otherwise."