Using deep learning models to improve climate change projections
The below case study shares some of the technical details and outcomes of the scientific and HPC-focused programming support provided to a research project through NeSI’s Consultancy Service.
This service supports projects across a range of domains, with an aim to lift researchers’ productivity, efficiency, and skills in research computing. If you are interested to learn more or apply for Consultancy support, visit our Consultancy Service page.
Research background
New Zealand needs to adapt to and mitigate the effects of climate change to build a climate-resilient economy. To deliver the full information needed for societal decision-making, high spatial resolution climate projections are required.
General Circulation Models (GCMs) are sophisticated physics-based models for simulating the response of global climate to increasing greenhouse gas emissions. Current GCMs are extremely computationally expensive which makes running high spatial resolution ensembles currently unfeasible – even on the world’s largest supercomputers. The resolution of GCMs are usually in the order of 100km, which is too coarse to deliver the full information needed for societal decision-making at regional and local scales.
Existing methods to enhance the resolution of climate projections include dynamical downscaling, which is also very computationally expensive. To dramatically reduce the computational burden of dynamical downscaling, NIWA Data Scientist Neelesh Rampal alongside with Climate Scientists Peter Gibson and Nicolas Fauchereau developed a state-of-the-art deep learning model to leverage high-resolution rainfall observations produced by NIWA. Our deep learning model will enable accurate, accessible, and timely climate change projections to assist the management of mid to long-term climate-related risk to New Zealand’s economy. Neelesh is also working alongside Meteorologists Ben Noll and Tristan Meyers, to provide high spatial resolution sub-seasonal to seasonal drought forecasts.
Applying this state-of-the-art deep learning model on the extremely large archives of climate projections (petabytes) requires scaling-up the training and inference steps as well as automation of the whole pipeline.
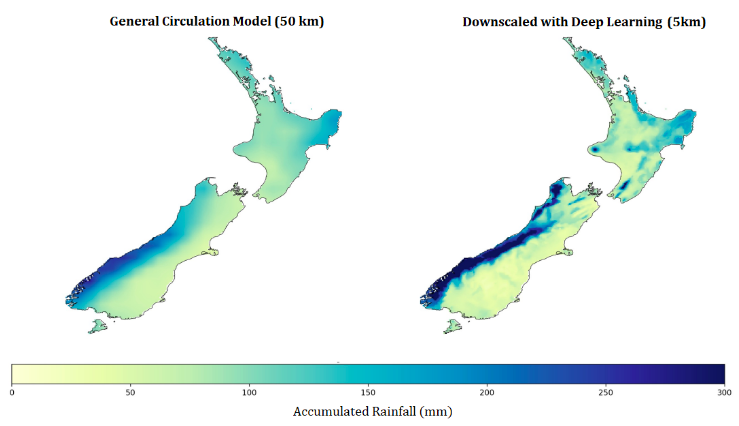
The Virtual Climate Station Network (VCSN) data contains observation data of NIWA climate stations interpolated to a 5km grid covering New Zealand. In order to improve the accuracy of the interpolated data, NIWA Data Scientist Neelesh Rampal developed a deep learning model to leverage simulations produced by NIWA. Applying this new technique on the full dataset requires scaling-up the training and inference steps as well as automation of the whole pipeline.
Project challenges
Neelesh needed help to deploy his code on NeSI's HPC platform, to train deep learning models on GPUs in an automated and seamless manner. Furthermore, one of the significant limitations of Neelesh’s existing workflow is that he wasn’t able to scale-up the amount of data he could train models on while limiting the amount of memory needed.
What was done
NeSI Data Science Engineer Maxime Rio and Application Support Specialist Wes Harrell helped Neelesh get started on NeSI’s HPC platform. This included showing him the ropes and basic 'how-tos', as well as providing particular guidance on how to automate Neelesh' workflow using Snakemake, a workflow management system, in combination with Papermill, a tool to execute Jupyter notebooks non-interactively.
In addition, Maxime developed a specialized Python class to enable loading data directly from disk. This piece of code enabled a reduction in the amount of memory needed while keeping training time within reasonable time bounds, using multiple workers to load and prepare mini-batches of data in parallel with the model training loop. On a synthetic task involving fine-tuning a ResNet-50 model on 50 GB of data, the new loader consumed only 15 GB of memory with a slowdown of 1.2X compared to preloading all data into memory before model training.
Main outcomes
Implemented a data loader enabling training models on very large datasets, loading directly from disk while keeping a low overhead.
Neelesh gained new skills as a researcher, becoming proficient at using the Mahuika cluster for his deep learning models training.
Researcher feedback
"The NeSI consultancy which was led by Maxime (Rio) and Wes (Harrell) has enabled me to significantly advance my abilities to design and automate workflows on the NeSI HPC platform.
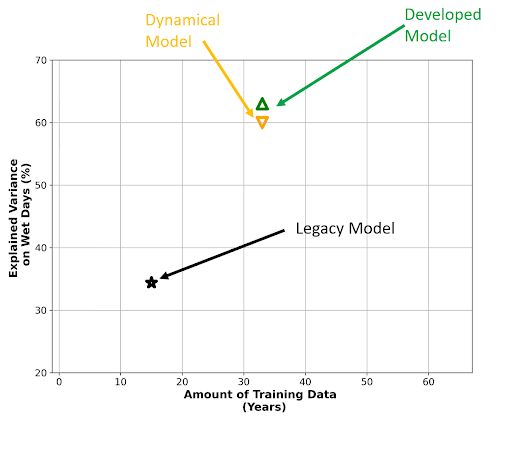
The advantage of learning how to create workflows on the platform, is that we could create reproducible science which enables better collaboration amongst scientists. The most significant outcome of the NeSI consultancy was the development of a Python package that enabled me to train deep-learning models on extremely large climate datasets, which before were not possible due to memory issues. Because our deep learning models can learn from larger datasets, we are able to achieve better model performance."
- Neelesh Rampal, NIWA Data Scientist
Do you have a research project that could benefit from working with NeSI research software engineers or our data engineer? Learn more about what kind of support they can offer and get in touch by emailing support@nesi.org.nz.







