The University of Otago is at the technological frontier
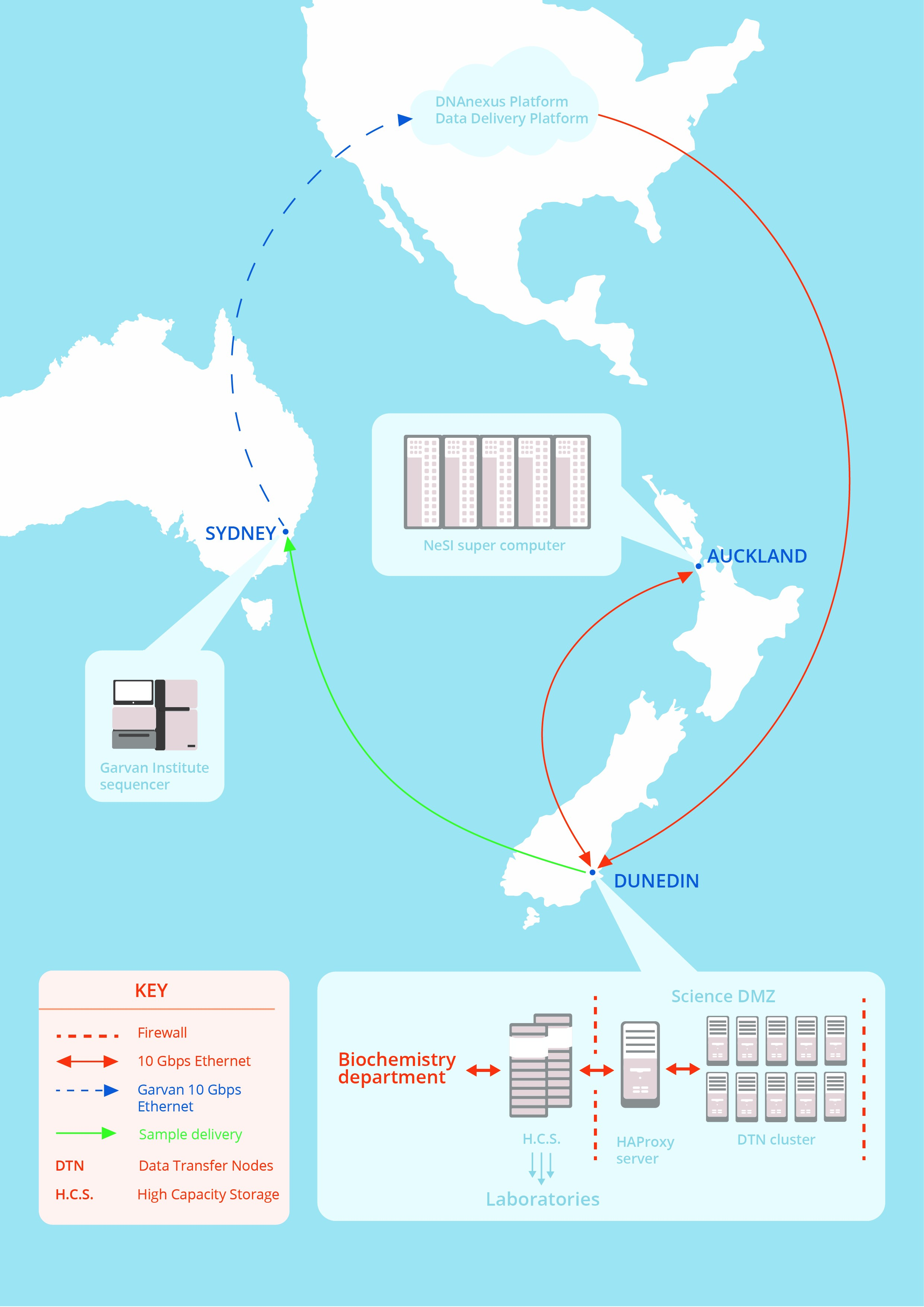
University of Otago researchers can now send and receive enormous amounts of data at ultra-fast speeds to stay at the forefront of their field – prompted by Dunedin gout researchers wanting to develop IT capability in New Zealand.
The researchers had to get up to 100 terabytes of data from the United States, send it in chunks for processing at a NeSI supercomputer in Auckland, then get the results back to Dunedin, says David Maclaurin – University of Otago Information Technology Services (ITS) Systems Services Manager, and NeSI Site Manager.
Ground-breaking research
He and his ITS team went “a very pale shade” as Biochemistry Professor Tony Merriman explained the IT support needed for his gout research, which could spark new treatments to prevent and treat the painful form of inflammatory arthritis.
The condition involves too much uric acid ending up in people’s blood instead of their urine and forming needle-like crystals in their joints, which sparks stiffness, swelling, heat, and pain.
Gout is most common in Pacific countries and some ethnic groups – such as Maori and Pacific people – are particularly susceptible to it. But developed countries such as New Zealand also have a high burden of gout and that burden seems to be becoming heavier.
All about gout
Professor Merriman wants to confirm the results of smaller studies about which genes underlie gout, while also isolating new genes that could raise the risk of developing gout, influence how serious the condition gets, and be a factor in flares.
The researchers are also analysing any associations with genes responsible for other illnesses that often affect people with gout - including heart disease, type 2 diabetes and kidney problems - as these conditions could themselves be part of the cause of gout.
“An important tool in this research is the ability to determine the DNA sequence of the inherited genome … this project represents significant technical challenges in data transfer and analysis.” Professor Tony Merriman, Department of Biochemistry, University of Otago
A big task
The project started with Otago researchers sending DNA samples of 1250 genomes – a complete set of genes from 1250 people with gout worldwide – to the Garvan Institute of Medical Research in Australia for sequencing.
A human genome – found in any cell with a nucleus – contains more than three billion DNA building blocks, known as bases, and the Otago researchers need the sequencing to reveal the order those bases appeared in each whole genome.
Then, the researchers can compare individuals and look for differences relevant to gout, on a scale never attempted before.
The terror of terabytes
Mr Maclaurin says the data pipeline is so complex, “we spent hours with whiteboards, trying to work out how it should all work and how it should look. But with a lot of good people contributing, it becomes possible.”
Over the project’s lifetime, about half a petabyte of data will be transferred and stored.
Tackling security and bandwidth
The ITS team’s first challenge was to get the 100 terabytes of raw, anonymized, fragmented, sequencing data to Dunedin from a global cloud-based genomic data delivery platform in the United States called DNAnexus.
Fortunately, the team was already researching and installing technology to get data both into and out of the campus more quickly.
The team was working with the Research and Education Advanced Network New Zealand (REANNZ) installing hardware known as a managed network edge and configuring several types of software, to create a suite of services known as a Science DMZ, which is a demilitarised zone between networks that is specifically designed to allow big research data sets through with the fewest possible obstructions, at the highest speeds possible.
The collaborators ended up with everyday University internet traffic still arriving and leaving on pathways with the usual checks, while authorised research data by-passed any checks it did not need.
Creating a data transfer engine
The ITS team was also already collaborating with NeSI staff at NIWA – the National Institute of Water and Atmospheric Research – on a blueprint for an ‘engine’ that could drive the transfer of large volumes of data at continuously high speeds.
The collaborators were using Globus based Data Transfer Node software, that had not been combined with a Science DMZ in New Zealand before, to create a Data Transfer Service for the University of Otago.
The Otago team had gone down both those paths because it already had another major research IT project underway.
Two game-changers
“We were now juggling the IT requirements for two large, complex and game-changing research projects simultaneously,” Mr Maclaurin says.
“We spent a year literally going from one meeting to another saying ‘that will be fine, that will be fine’ then walking out of the room and saying ‘right - how do we design this bit and get this going, what do we need to build first, and who do we need to collaborate with?"
“But we weren’t going to let it fail. We were going to make sure we got there in some way, shape, or form.”
Knocking it all into shape
The ITS team also soon realised many data pipeline components would have to be scaled up or built from scratch because of the project’s scale.
But collaboration with NeSI and REANNZ let the team talk to other people with similar challenges and re-use and adapt some of their ideas.
Project milestones included:
- Spending many months at the whiteboard designing components, testing ideas, and calculating the necessary data throughput and capacity.
- Adding more high-capacity Isilon storage units to the University’s High Capacity Storage clusters that can hold petabytes of research data.
- High-speed network links extended into the Department of Biochemistry from the University’s Data Centres to support the data pipeline.
- Commissioning the Science DMZ and repurposing two chassis of compute blades - from computers at the end of their lives – to create a new 10 node NeSI-Otago data transfer cluster, split between the University’s two Data Centres. The ITS team calculated that this could drive the data transfer throughput required.
- Successfully building proxy servers to ensure that the research storage clusters could communicate securely with the Data Transfer Nodes.
- Reviewing the whole environment and service procedures several times to minimise security risks.
- Load testing, speed testing, and tuning, which took four months.
- Creating procedures, manuals, and trouble-shooting guides for University-wide use.
Supercomputer chunks
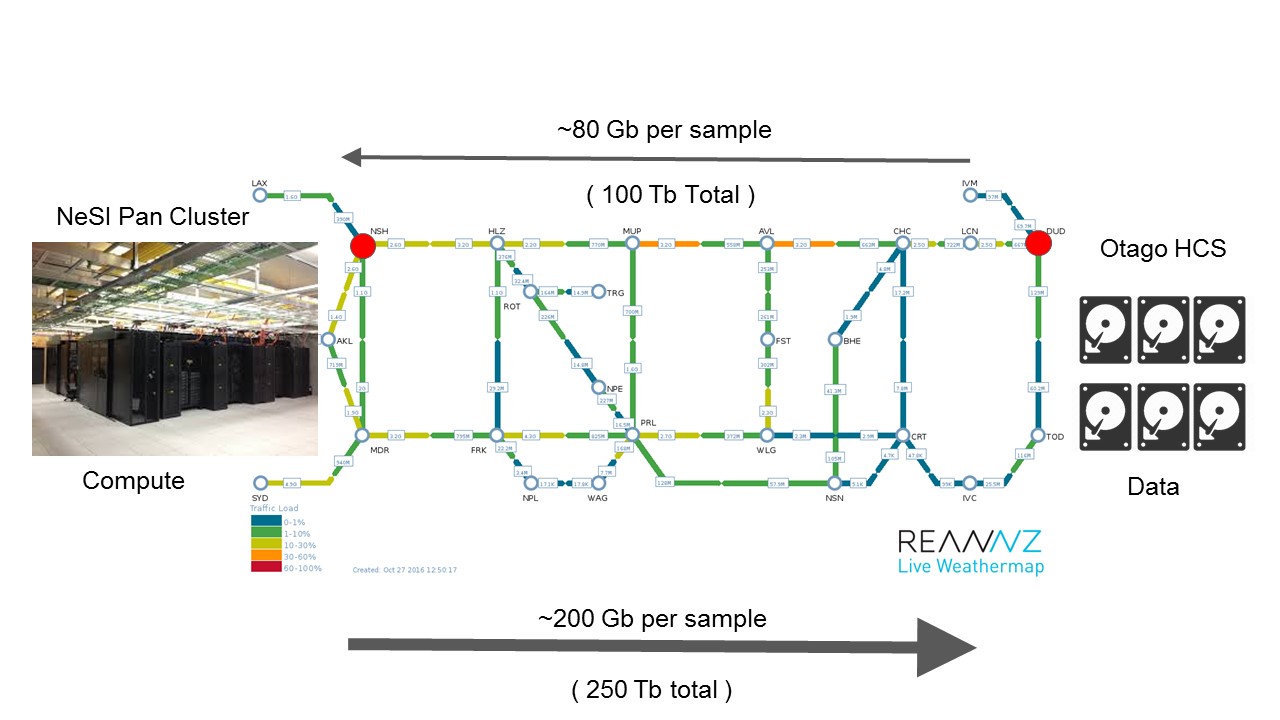
But the ITS team still had to get the raw sequencing data from Dunedin to the NeSI PAN supercomputer in Auckland.
Because the supercomputer has 50 terabytes of RAM, about 6000 Central Processing Unit cores, and half a petabyte of disk – compared to a standard laptop’s four cores and eight gigabyte RAM – it can run hundreds of research jobs simultaneously every day. But the Otago project is so large, its data has to be processed in chunks so the supercomputer can keep processing other jobs as well.
So, Otago postgraduate biochemistry student Murray Cadzow wrote a program which calls the University’s new Data Transfer Service to get it to send 80 gigabyte slices of data north on the superhighway provided by REANNZ, then gets the supercomputer to start processing the data.
Parallelising work
His coding added a Genome Analysis Toolkit – created by the Broad Institute in the United States – to the supercomputer as well.
His coding also splits the 80 gigabytes into smaller slices so they can all run through the first two stages in the toolkit in parallel – to align and sort the data – and are processed much faster.
Another break-up
His program then merges the smaller files again, before breaking them up a second time to process the data for each chromosome in parallel.
By then, the processing has identified where each DNA sequence sits in each whole genome in the file, and has summarised the differences for each sample.
Multiple files
Mr Cadzow’s program also allows multiple 80 gigabyte files to be processed simultaneously and polls the supercomputer so it tells him when the processing has finished, then sparks another data transfer to send the results to Otago.
If anything goes wrong, only a slice of data has to be handled again rather than huge amounts.
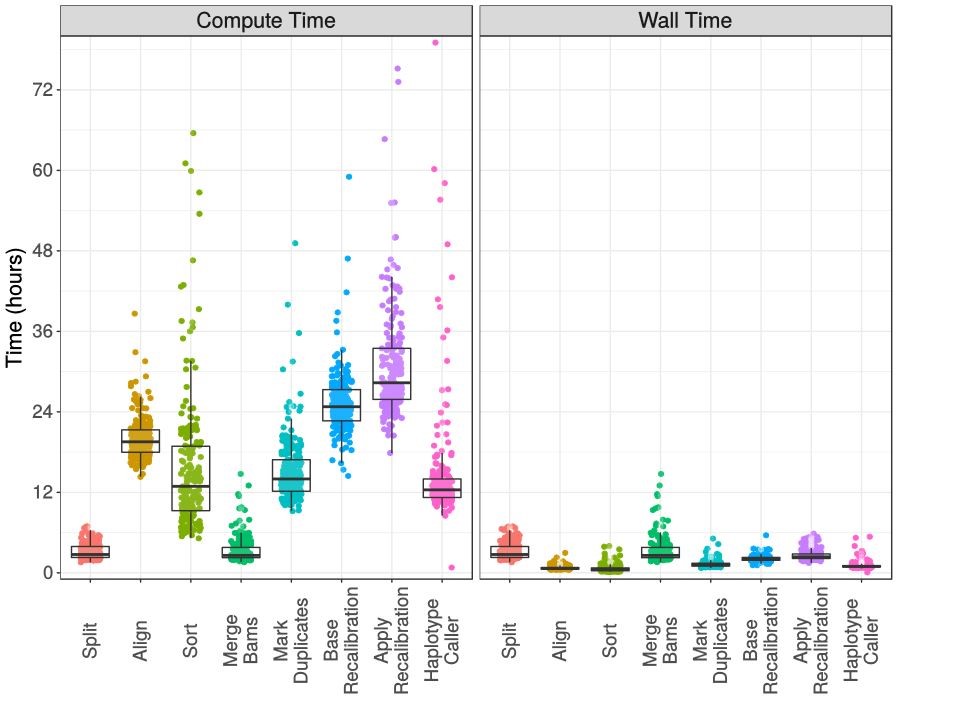
The processing steps that strongly benefit by parallelisation, are evidenced by highly variable compute times on the left (the vertical coloured data point streaks), but relatively uniform elapsed times on the right. The x-axis itemises the genomics data processing steps being undertaken on the NeSI supercomputer.
Getting out of the way
While Mr Cadzow had support from Mr Maclaurin’s team and NeSI to make sure his data pipeline ran smoothly, he was mostly left to his own devices because “he understands both the technology stack and the academic domain”, Mr Maclaurin says.
“He knows what he’s trying to achieve with the research, but we don’t. I’m happy Murray has written his own code because it’s exactly what genomics people need and he’s now sharing that data pipeline code with other researchers around New Zealand.”

“We get out of the way when we need to get out of the way, but we help when our expertise is needed.”
The supercomputer will now process gout data for the next 12 months – day and night – to return about 250 terabytes of data to the University in total.
"Without the help of Otago ITS and NeSI … this project would not have been possible to do within New Zealand." Murray Cadzow, University of Otago Biochemistry Department postgraduate student.
The future
Professor Merriman and his group will be busy processing and analysing the data over the coming year. Mr Maclaurin, his team and NeSI will still be working closely with them: “We are committed to ensuring that their research is successful and completed on time. If problems arise, we step in as quickly as we can.”








