Understanding proteins through simulation
The problem:
Some species of bacteria can be dangerous or deadly to humans. Discovering new drugs to fight these bacteria without hurting the hosts is difficult work. An enzyme called MenD is vital to many species of bacteria but not humans. A natural regulation site on MenD was identified but it’s unclear how it works.The solution:
With the help of molecular dynamics simulations, researchers can better understand how MenD works and its mechanism of regulation. Knowledge gained could be used to design new drugs to fight the bacteria causing diseases such as tuberculosis.The outcome:
The mechanism of regulation on MenD enzyme was identified. Researchers will now try to validate the predictions through experiments in the lab.
Dr Wanting Jiao is a computational biophysicist and biochemist at the Ferrier Research Institute, Victoria University of Wellington. There, she models proteins to understand how they behave under different conditions. Wanting has worked with NeSI for over a decade. The long-term partnership has helped her model enzymes and provide important insights for the design of new antibacterial medicines.
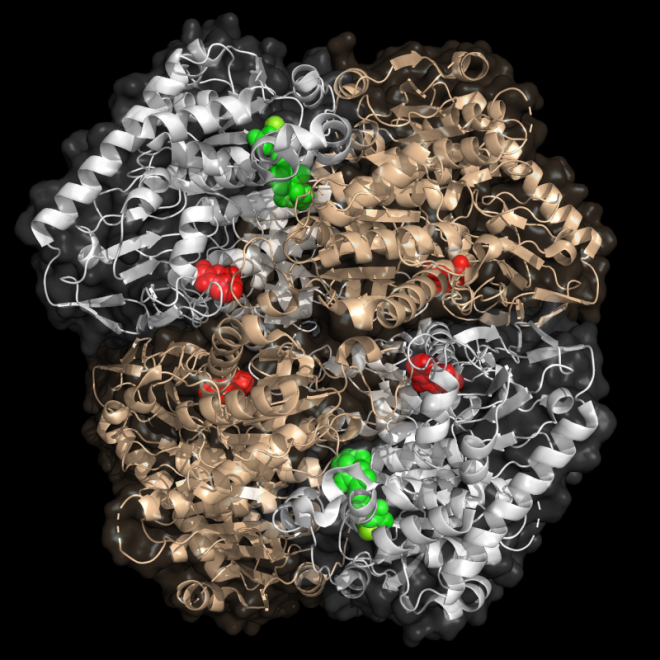
Wanting used NeSI’s Maui supercomputer to model the MenD enzyme. MenD is the first enzyme in the biosynthetic pathway that produces Vitamin K2 in many bacteria. Humans get their Vitamin K2 from foods, so we don’t rely on this enzyme. But MenD is vital for many species of bacteria, including Mycobacterium tuberculosis, responsible for tuberculosis in humans.
“MenD is essential for the survival of many pathogenic bacteria, so it’s a good target for making antimicrobial drugs. If we can understand how the enzyme works we can stop it and kill the bacteria,” Wanting said.
But MenD is a large enzyme and the simulated system contains over 20,000 atoms. Each atom is explicitly modelled. This makes MenD impossible to simulate on most computers.
“We have more than 20,000 atoms in this modelled system and there are four different states I need to study. For each state I need to do replicates, so the number of calculations increases very quickly,” Wanting said.
“We can’t run this program on our lab computers. Even small clusters, like a 12-CPU Linux, are not powerful enough to simulate MenD. That’s why the NeSI platform is crucial to what we’re trying to do.”
Enzymes like MenD have an active site. This is the area where the enzyme binds its substrates, to carry out the job it is designed to do. Many drugs target enzyme active sites because interfering with them works like a stop order for the enzyme’s job. However, MenD is naturally regulated by another mechanism.
“MenD has an active site, but it’s regulated by a small molecule binding to a distant area of the enzyme. It’s a kind of remote control. We want to understand how the signal propagates through the protein structure using our simulation.”
This control site could be the key to kill bacteria that use the enzyme. If researchers can synthesise a molecule to use on the control site, it could be a safer, more targeted solution to stopping bacterial growth.
Wanting was given one million core hours on NeSI’s Maui supercomputer for her project. Even with this time, the data was so complex Wanting knew every minute would be needed. The simulation code would also need to be as efficient as possible. To ensure this, Wanting worked with NeSI research software engineer, Chris Scott, to optimise the code.
“Chris helped with a range of things: he helped optimise our automatic submissions script and made the simulation more efficient. Chris recompiled the simulation code, resulting in a 37 per cent reduction in run time. He was always willing to listen to my issues and provide help when problems arose,” Wanting said.
The next stage of Wanting’s research is to take her discoveries back to collaborators, a team of scientists across New Zealand led by Dr Jodie Johnston at University of Canterbury, to see if they can validate the predictions in a lab setting.
“This project isn’t just about developing new medicines, we’ve also been able to add to the fundamental knowledge of how enzymes work. By mapping the atom movements as they propagate through the enzyme, we get an idea of how the organism regulates the enzyme. Researchers could use this knowledge when looking at other enzymes in the future,” Wanting said.
For now, Wanting’s research is geared towards disrupting the MenD enzyme. The simulations will provide new understanding of how MenD works that otherwise may be hard to access using experimental approaches. It would not be possible without the expert advice and the supercomputing resources of NeSI.
Do you have an example of how NeSI support or platforms have supported your work? We’re always looking for projects to feature as a case study. Get in touch by emailing support@nesi.org.nz.