Tools to better understand and address water quality issues
The below case study shares some of the technical details and outcomes of the scientific and HPC-focused programming support provided to a research project through NeSI’s Consultancy Service.
This service supports projects across a range of domains, with an aim to lift researchers’ productivity, efficiency, and skills in research computing. If you are interested to learn more or apply for Consultancy support, visit our Consultancy Service page.
Research background
Thanh Dang, Sandy Elliott and Linh Hoang are scientists at NIWA who apply mathematical techniques to address water quality issues. One of the applications of this work is to predict the effects of diffuse pollution and its control at catchment scale.
Thanh has developed an R code within the R-SWAT framework to perform calibration, sensitivity and uncertainty analysis of Soil Water Assessment Tool (SWAT) runs.
For large catchment areas and/or large numbers of parameter runs, SWAT execution can take days on a personal computer. Therefore there is a need to migrate such parameter studies to a high performance computing platform that can leverage hundreds or more cores to reduce the turnaround time.
Project challenges
The researchers wanted to set up a framework that is capable of leveraging hundreds of cores to run parallel jobs. They approached NeSI for help with this task and worked with Research Software Engineers Alexander Pletzer and Chris Scott.
What was done
- investigated different approaches to scale an R cluster to more than one node
- determined that with a few coding changes, an MPI cluster that dynamically spawns MPI processes can be built and launched on NeSI platforms
- tested and recorded the parallel scalability of different parallelisation approaches and settings
Main outcomes
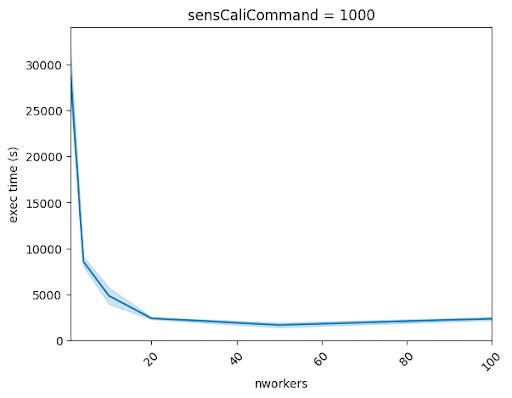
Best results were obtained using the mpirun -bootstrap ssh method on Mahuika Broadwell to start an MPI dynamic processes cluster. The figure below shows an example of execution time reduction for 1000 independent simulations, using a different number of “workers”. The parallel speedup is 22x in this case.
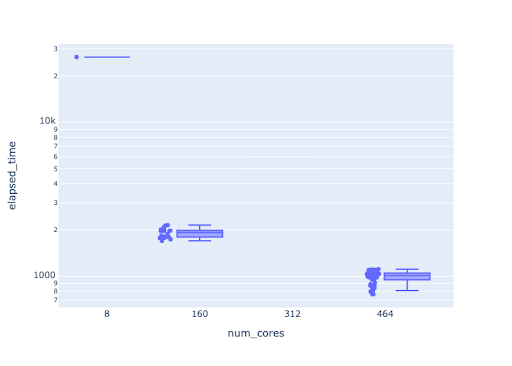
Another strategy, shown in the figure below, involves breaking the code into small executables which are run through a workflow, and assembling the final result. Note the log scale. In this example, the execution time was reduced from > 3 days (serial) to 20 minutes (50 worker x 8 threads).
Researcher feedback
"The team at NeSI worked with us to provide a solution to achieve significant speed-up in legacy R- and FORTRAN-based code for catchment model runs. They explored and trialled alternative solutions, including some tricky MPI switches, and documented run-time results including variability. They contributed to co-authored conference presentations, and came up with some new ideas to try in the future."
- Dr Sandy Elliott, Principal Scientist - Catchment Processes, NIWA
Do you want to bring your research to the next level? We can help. Send an email to support@nesi.org.nz to learn more about our Consultancy support.







