Tech Insights: Testing the A100 tensor cores on deep learning code
This is the second post in our blog series sharing some of NeSI's behind-the-scenes work to prepare our new A100 Graphics Processing Unit (GPU) cards for machine learning use. The previous article unveiled the installation, thermal stress test and internal testing.
This post in our Tech Insights series focuses on tests of deep learning code we did in partnership with our test users. We were particularly excited to try these types of applications so that we could test the A100s' tensor cores. These are a new type of computing unit especially made to accelerate the matrix operations found in deep neural networks, with more parallelism and the possibility to use mixed precision computations to increase its throughput.
DeepLogic, differentiable logical reasoning
The first two pieces of software we will talk about about were tested in collaboration with Qiming Bao, a PhD student in Michael’s Witbrock lab at the University of Auckland. Qiming studies how deep neural networks can be used to learn symbolic logic directly from textual inputs.
Our first contender, DeepLogic, is a neural network built using TensorFlow. The first challenge here was to install the right version of TensorFlow with its dependencies. On NeSI, you have multiple ways to run TensorFlow: using our pre-packaged Python module, installing it in a Python or Conda virtual environment, or running it in a Singularity container.
As we tried the different methods, we realised a couple of things:
- Some pre-packaged versions of TensorFlow 1 require an old version of Glibc (2.27), which is not available on NeSI's high performance computing (HPC) platform, so if you do need them, you will need to use a container.
- TensorFlow seems good at backward compatibility, within the major versions, so always try the latest version which will more likely have supported dependencies.
- Keep the TensorFlow build matrix at hand, to pick the right versions of Python, TensorFlow, CUDA and cuDNN.
Once we finally managed to get a functional environment, using TensorFlow 1.15, we ran DeepLogic on the A100 and… it crashed. Repeatedly.
It turned out that TensorFlow 1 does not provide support for the Ampere architecture, the architecture of the A100!
Fortunately, NVIDIA provides a collection of containers for popular tools and packages, including TensorFlow 1. They maintain a version compatible with their latest hardware. These are Docker containers, so all we had to do was convert them into Singularity containers, make sure that the container can use the GPU, and voilà!
Note: Singularity is a container technology well suited for an HPC environment, as it doesn’t require root access to run the container. Conversion from Docker images is straightforward.
However, this was not the end of our struggle with this piece of code. Comparing speed with the P100, running on the A100 did not bring any speedup, which is quite disappointing.
The good news is that the NVIDIA container is packed with goodies to address this point. One of them is the Nsight Systems, a profiler particularly good for profiling CPU-GPU interactions. NVIDIA even provides a wrapper, DLprof, to make it easier to profile deep learning code, capturing special events from deep learning toolboxes alongside data and kernel information.
To use it, one just needs to prefix the python command with dlprof and that’s it, easy peasy! Profiler results can then be explored using the nsys-ui graphical tool, already available on NeSI as a module and easily accessible via virtual desktops. Sadly, the results of the profiling did not reveal any obvious pathological pattern, e.g. a communication bottleneck between CPU and GPU.
In addition, DLprof also provides recommendations. In our case, it recommended us to use XLA optimization and turn on Automatic Mixed Precision to run some computations as FP16 on the tensor cores (as simple as setting an environment variable). Applying these changes did not improve the speed, so as of now the mystery of the bad performance is complete but investigation is still ongoing.
Fairseq, sequence modeling toolbox
Another tool Qiming wanted to evaluate is Fairseq, a toolbox for sequence modeling, published by Facebook and using PyTorch as backend. Our test task was a fine-tuning task of the RoBERTa language model.
After the bumpy ride with DeepLogic and TensorFlow 1, running Fairseq felt like a leisure cruise. First, installing PyTorch using pip just works™, one just needs to be careful to use the version supporting CUDA 11.1 to make it work on the A100.
Next, the toolbox works great out-of-the-box and exposes many options for advanced training (FP16, multiple GPUs and multiple nodes training) that will reveal themselves to be useful (as we will see soon).
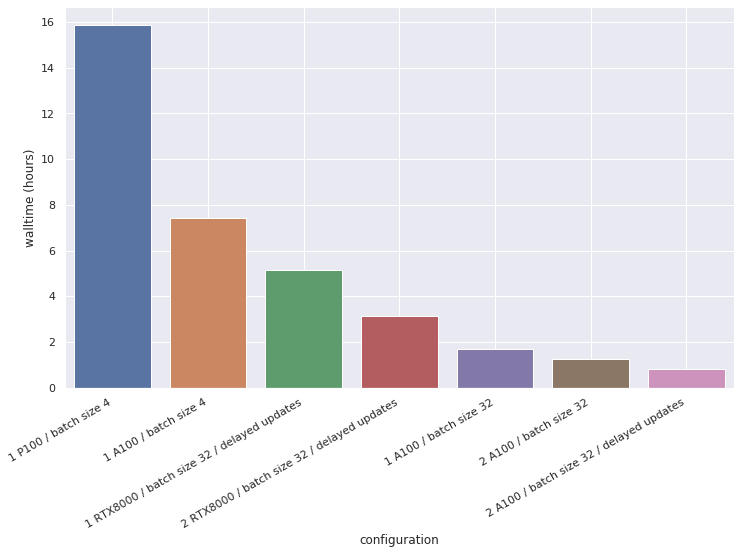
To compare runtime performances, we decided to monitor how much time was needed to run the fine-tuning for 10 epochs, without early stopping. The RoBERTa model was quite large for our P100 card, which has 12GB of memory, so the mini-batch size was limited to four.
Using the same mini-batch size, the A100 was 2X faster than the P100. But this scenario heavily under-used the A100 resources. With 40GB of memory, the A100 can handle much larger mini-batches. Using a mini-batch size of 32, the A100 ran the test code 10X faster than the P100.
The next logical step was to train on multiple GPUs. In our current installation of the A100 cards, there are at most two cards per node, without NVLINK bridge (see our previous post). Our first attempt at training the model with two A100s revealed itself unconclusive: performances were the same with one or two GPUs. What was wrong?
It turns out that multi-GPU training, in its simplest form, consists of sending different mini-batches of data to the different GPUs, to compute gradients for the same parameters of the model duplicated on each device.
Once these gradients have been computed, they must be exchanged between the GPUs to update the model parameters. The model being large, so are the gradients, which means that communication between the devices is crucial to keep training time low. This is what was hitting us here, the communication cost for the updating the model parameters after each mini-batch. One solution was to get a better bandwidth between cards, using an NVLINK bridge for example, which we don’t have unfortunately.
Another clever solution was to use delayed updates (see Ott et al. (2018) for more details). The trick is to accumulate gradients from multiple mini-batches on the same devices and transfer gradients only once every few mini-batches. The model is updated less often, with larger mini-batches and the cost of communication is amortized over multiple mini-batches. The figure below shows the delayed updates technique.
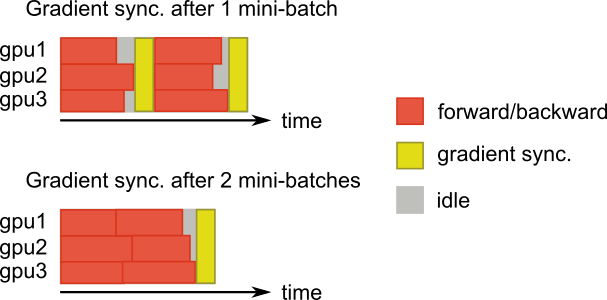
Using two A100 GPUs, a mini-batch size of 32 and updates every eight mini-batches, we managed to get a 20X speedup compared with one P100 GPU. In this scenario, the model is updated every 2 x 32 x 8 = 512 samples. This is one limitation of the approach: the mini-batch size is artificial increased, potentially above what what is sufficient to obtain a stable convergence of the model.
To complete this benchmark, Qiming also ran the same code on RTX8000 GPUs available in Michael Witbrock’s lab computers. These GPUs have more memory than our P100s, allowing for mini-batches of 32 samples. In a scenario with two GPUs and using delayed updates, the A100s were 4X faster than the RTX8000s. The following graph summarizes the tested configurations.
DeepLabCut, markerless pose estimation
A final example we wanted to share with you in this post is DeepLabCut. This software is used to estimate animal pose from videos, without any marker. It leverages pretrained deep neural networks, requires a moderate amount of data (~100 annotated frames), and uses transfer learning to train the model.
The resulting trained model can then do reliable tracking on hours of recordings. This work was done in partnership with Dr. Hamid Abbasi from the Auckland Bioengineering Institute at the University of Auckland.
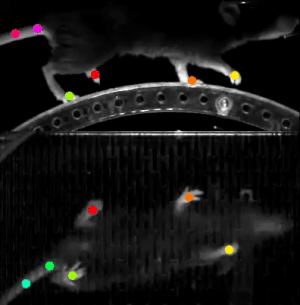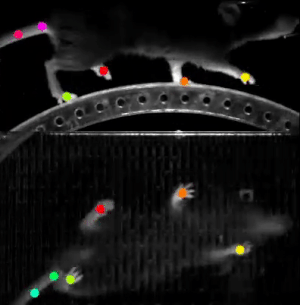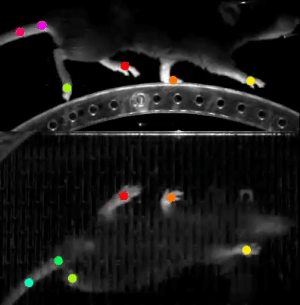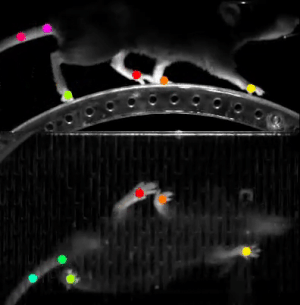
This software relies on TensorFlow 1 (a version using TensorFlow 2 is on its way). We learned our lesson here and created a custom Singularity container, based on the NVIDIA TensorFlow 1 container. It just worked.
Compared to the runtime on a P100 card, fitting a ResNet-152 model was 2.3X faster on an A100. More importantly, the larger memory of the A100 allowed Hamid to select bigger models, such as EfficientNet-B6, as the base model for transfer learning. This model would just not fit on the P100 card.
Conclusion
In this second part to our A100 Tech Insights series, we presented a sample of our users' tests using the A100 GPUs for deep learning. Testing real user code, instead of benchmarks, gave us the opportunity to learn more about the experience we can expect for our users and ways to make it as smooth as possible.
Thus, we learned that it is best to recommend the NVIDIA container for any TensorFlow 1 application. Anything else was doomed to fail to run on the A100. For PyTorch and TensorFlow 2, using the latest version supporting CUDA 11.1+ is enough.
The other interesting point to emphasize is the importance of the GPU memory. Without any modification, applications were running at least 2X faster on a A100 compared to a P100. But more interestingly, some pieces of code couldn’t run at all without enough GPU memory, due to model size, or would be very limited by a small mini-batch size.
In the next instalment of this series, we will focus on another area where GPUs shine, molecular dynamics code.
If you are interested in learning more about these new GPU resources and whether they would be of use to your project, please contact NeSI Support.








