Fishing for parallelisation strategies
The below case study shares some of the technical details and outcomes of the scientific and HPC-focused programming support provided to a research project through NeSI’s Consultancy Service.
This service supports projects across a range of domains, with an aim to lift researchers’ productivity, efficiency, and skills in research computing. If you are interested to learn more or apply for Consultancy support, visit our Consultancy Service page.
Research background
To explore different fishing scenarios under different environmental conditions (e.g. climate change), one needs a numerical tool that integrates spatiotemporal, multi-population fish population dynamics models with statistically fitted observation data.
The SEAPODYM modelling framework allows building quantitative spatiotemporal models for migratory fish species with different life history traits, by integrating observations to inform model parameters. Inna Senina is a Senior Fisheries Scientist at The Pacific Community (SPC) whose work includes modelling tuna population dynamics. She uses the SEAPODYM framework, however, the runtimes for parameter estimations can be extremely long, limiting her reference model configurations to coarse resolutions and/or short simulation times.
The objective of this Consultancy project was to review Inna's SEAPODYM computer code to provide potential solution(s) for its parallel execution.
Project challenges
To identify potential parallelisation strategies, NeSI Research Software Engineers Alex Pletzer and Chris Scott needed to acquire some understanding of how SEAPODYM works, in particular how different populations of fish (cohorts) are grouped by age. The latter affects the longevity of the group’s members and how many time steps should be taken in the future before most members are removed from the system.
Each cohort starts with initial conditions in the simulation, which involve accumulating the density of other cohorts at the previous time step. A particular challenge was to design a workload for parallel workers that produced good load balancing (i.e. workers won’t have to wait for other workers to finish their task).
What was done
Alex and Chris identified numerically expensive loops which can be vectorised or parallelised with OpenMP directives. Next, they looked at MPI parallelisation and developed a toy model, which integrates in time the equations describing cohort dynamics (cohort - a group of individuals born at the same time), replacing the advection-diffusion-equation with a sleep instruction.
A second part of the project involved investigating whether the Alternating-Direction Implicit (ADI) time advance could be replaced with an explicit time stepping method, which could be parallelised on its own right.
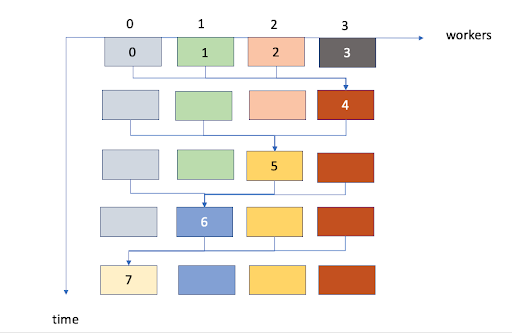
Figure: MPI parallelisation over cohorts (i.e. group of fish that were born the same year). Each colour represents a different cohort. In this case cohorts 0, 1, 2 and 3 can run the first step in parallel. At the next time step, a new cohort (4) is born, which requires data from cohorts 0, 1 and 2. Note that if each block takes the same execution time then there is perfect load balancing.
Main outcomes
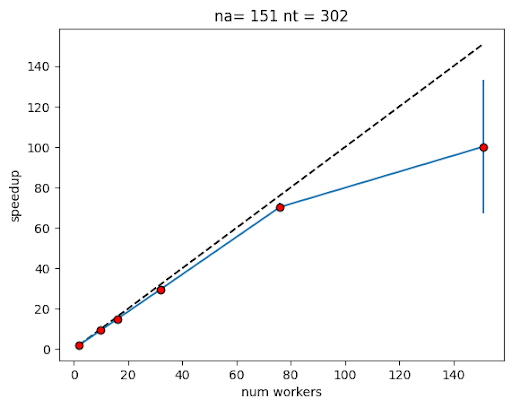
Based on estimates for the time it takes to advance the system by one step for one cohort and the amount of data that needs to be exchanged after each step, NeSI found that a speedup of up to 70x could be achieved for the larger, forward problem (for species with longer life span, e.g., albacore, bluefin) when using 76 workers.
A stability condition for the maximum time step based on the velocity and diffusivity was derived for the 1D case. It was found that when the minimiser integrates the equations backwards to estimate the gradient of the likelihood function with respect to the parameters, a fully implicit time stepping method should be used to recover the original initial condition.
Researcher feedback
"Alex and Chris were very insightful and productive, making in-depth revision of the code and suggesting plausible parallelization strategies that include different methods, and demonstrating how they work either with the actual code, or with a toy model. They were also very keen to help with other questions and issues, including code profiling, GitHub actions, OpenMP performance, etc. This work is the first phase of the SEAPODYM parallelization project, and given the very encouraging results and perspectives, I will be looking forward to continuing our collaboration with Alex and Chris on the second phase and implementing together the new parallel version of the SEAPODYM model code."
- Inna Senina, Senior Fisheries Scientist, The Pacific Community
Do you want to bring your research to the next level? We can help. Send an email to support@nesi.org.nz to learn more about our Consultancy support.







